Whilst most molecular HLA typing methodologies remain relatively static, next-generation sequencing (NGS) has tended to evolve quickly as product manufacturers respond to user suggestions, reagent development and changes in sequencing technology. One Lambda released AllType™ in Summer 2017: a NGS workflow for all loci, all platforms, all in a single reaction. This NGS workflow was a game changer, beginning with the long-range PCR of all 11 classical HLA genes in a single multiplexed reaction. Whilst AllType™ remains a competitive workflow, One Lambda have now released an alternative NGS solution: AllType™ FASTplex™.
AllType™ FASTplex™
The rationale behind AllType™ FASTplex™ was to simplify the NGS workflow, save time, and yet still generate high-quality results. FASTplex™ sets a new standard of simplicity for NGS and is a simple assay that anyone can learn. Like the classic AllType™ workflow, FASTplex™ begins with all 11 classical HLA loci (HLA-A,B,C,DRB1,DRB3/4/5,DQA1,DQB1,DPA1,DPB1) co-amplified in a single multiplexed reaction (Table 1). After an initial thermal cycling time of only 2.5 hours, patient-specific amplicons are pooled into a single tube within an hour of commencing library preparation. This means that approximately 70% of the FASTplex™ protocol is single tube workflow. The consequences of this are significant: from DNA to sequencing, sample preparation takes less than seven hours and requires only 90 minutes of hands-on time. An easy single-tube workflow reduces the chances of pipetting error and means that automation isn’t required. By eliminating most of the pipetting, purification and quantification steps required by ‘traditional’ methods, FASTplex™ significantly reduced the consumables required. Compared to the classic AllType™ workflow, FASTplex™ uses less than half the plastics, pipette tips and consumables: good news for both operational costs and the environment. The AllType™ FASTplex™ product also includes paramagnetic beads, further decreasing the cost per test resulting from consumables and third-party reagents.
Table 1: Gene content and coverage for AllType™ FASTplex™.
| Loci | Coverage |
| HLA-A | Whole gene |
| HLA-B | Whole gene |
| HLA-C | Whole gene |
| HLA-DRB1 | Exon 2 – Intron 5 |
| HLA-DRB345 | Exon 2 – Intron 5 |
| HLA-DQB1 | Exon 2 – 3’ UTR |
| HLA-DQA1 | Whole gene |
| HLA-DPB1 | Exon 2 – 3’ UTR |
| HLA-DPA1 | Whole gene |
The FASTplex™ Workflow
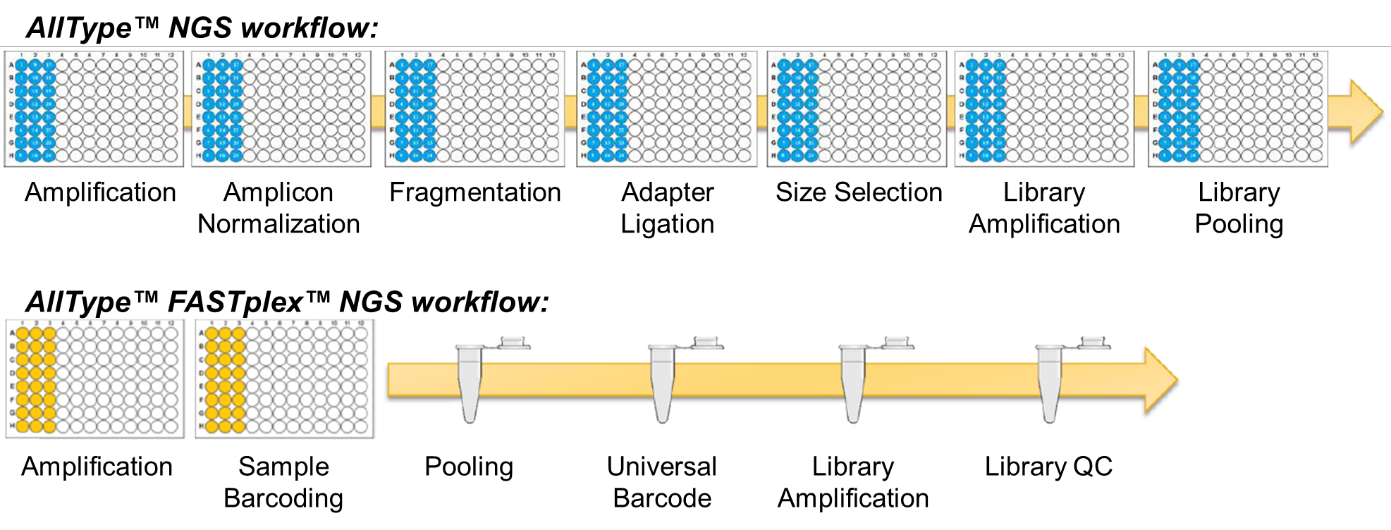
The AllType™ and AllType™ FASTplex™ workflows are compared in Figure 1. FASTplex™ builds on the classic workflow and removes some elements that might be considered undesirable in a non-automated laboratory. Following amplification and paramagenetic bead purification, sample-specific amplicons are quantified using a Qubit instrument. AllType™ requires sample-specific normalisation to proceed with 100 ng DNA whilst FASTplex™ allows the user to proceed with 3-30 ng DNA, reducing the pipetting burden as samples ought to fall within this range. A sophisticated workbook-based tool guides the user through the FASTplex™ workflow, indicating if adjustments are required and performing the necessary calculations. Unlike the classic workflow where fragmentation, adapter ligation (‘barcoding’) and size-selection are discrete processes, the FASTplex™ workflow simultaneously fragments and barcodes amplicons using a set of ‘FAST’ reagents pre-plated in a 96-well tray; size-selection is not required. At this point FASTplex™ samples are pooled and the remainder of the workflow occurs in a single tube. In the FASTplex™ workflow an additional ‘universal barcode’ is ligated to all fragments prior to a secondary PCR amplification. Thereafter the library is cleaned using paramagnetic beads, quantified and diluted to the concentration required for sequencing. Working in a single-tube significantly increases the speed and simplicity of the workflow; although bead-based purification and quantitation is still required the volume is significantly reduced compared to e.g. 24 samples.
Figure 1: The AllType™ and AllType™ FASTplex™ workflows compared.
Multi-Platform Support
Both AllType™ and AllType™ FASTplex™ are now supported on multiple Illumina platforms and kits, whilst AllType™ is also supported on various Ion Torrent platforms (Table 2; an Ion Torrent-compatible FASTplex™ workflow is pending). This provides verified flexibility for the end user, who may choose to utilise a sequencing platform shared by multiple users or acquire their own ‘entry level’ sequencer (e.g. the iSeq 100, with a purchase price of less than £20,000). Many sequencing platforms also offer different ‘kit’ sizes which allow users to implement a solution appropriate to their laboratory, i.e. balancing throughput, ‘batch’ cost per sample (considering the sequencing kit) and turnaround time.
Table 2: Sequencer throughput for AllType™ and AllType™ FASTplex™.
| Platform | Kit | Throughput | Sequencing Time |
| MiSeq | Standard | ≤ 96 samples | 24 hrs |
| Micro | ≤ 24 samples | 19 hrs | |
| Nano | ≤ 8 samples | 17 hrs | |
| MiniSeq | High-Output | ≤ 96 samples | 24 hrs |
| Mid-Output | ≤ 72 samples | 17 hrs | |
| iSeq 100 | i1 Reagent | ≤ 24 samples | 17.5 hrs |
| Ion GeneStudio S5 | 530 Chip | ≤ 48 samples | 21.5 hrs * |
| Ion GeneStudio S5 Plus | 530 Chip | ≤ 48 samples | 8 hrs * |
* FASTplex™ launch pending; does not include Ion Chef templating time.
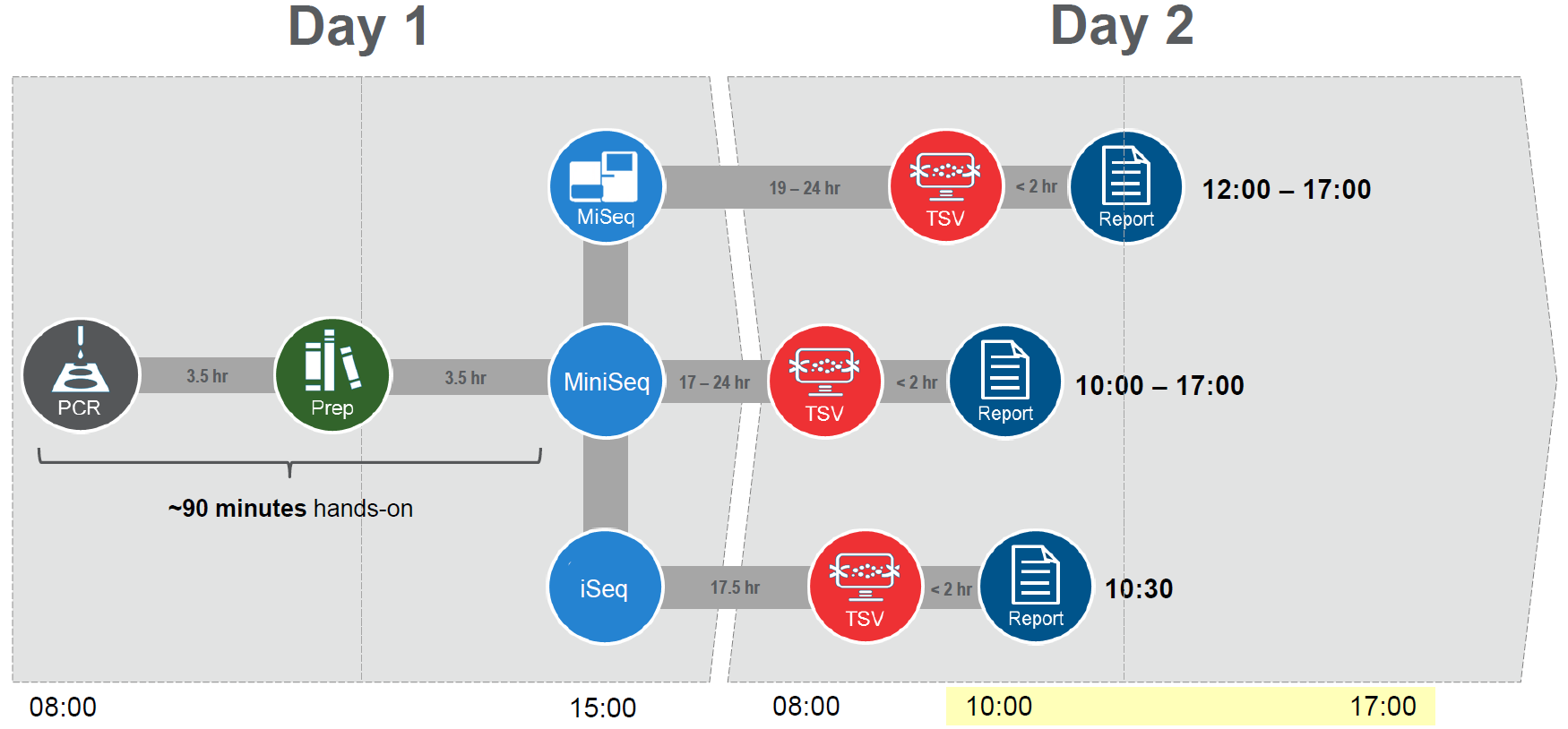
The sequencing platform and kit used also impacts the sequencing time and thus the time taken from sample to HLA genotyping results. Using the fastest solution, AllType™ FASTplex™ allows the user to obtain HLA genotyping results a mere 26 hours after starting their testing (Figure 2).
Figure 2: AllType™ FASTplex™ is the fastest sample-to-result NGS workflow for HLA.
TypeStream Visual™ 2.0: Fast, Fully-Automated Analysis
The release of AllType™ FASTplex™ coincides with the launch of TypeStream Visual™ 2.0: intelligent software for the analysis of AllType™ data from Ion Torrent, Illumina and PacBio sequencing platforms. TypeStream Visual ™ 2.0 retains the familiar (Fusion-esque) ‘front-end’ environment but builds on LIMS integration: SystemLink offer a HistoTrac module that interfaces directly with the TypeStream Visual™ database, and HLA typing data can now be exported to One Lambda’s HLA Fusion™ software with only three clicks. This allows the integration of high-resolution genotypes with patient/donor records and facilitates antibody tracking and epitope-level analysis using HLAMatchmaker functionality built into HLA Fusion™.
The TypeStream Visual™ auto-analysis feature streamlines data migration so that data are analysed even when the user is away from the laboratory. TypeStream Visual™ 2.0 features a new analysis engine that allows multiple samples to be analysed simultaneously, resulting in analysis times less than half that of TypeStream Visual™ 1.X. This ensures that AllType™ and AllType™ FASTplex™ are the fastest sample-to-result NGS workflows for HLA genotyping. TypeStream Visual™ 2.0 offers more user-definable features including health metrics, system comments and the ability to exclude specific gene regions (e.g. micro-satellites). The analysis experience has been redesigned with a streamlined interface: fewer ‘levels’ of analysis require fewer ‘clicks’ to assign a result (Figure 3). Upgrading to and maintaining TypeStream Visual™ 2.0 has also been made easier with a result comparison tool that can also be used to verify initial and confirmatory HLA typing data.
Figure 3: TypeStream Visual™ provides the user with a streamlined analysis experience.

NGS is complicated – but your workflow doesn’t have to be. Learn how you can adopt the AllType™ FASTplex™ method for HLA typing that’s simple, fast and best-in-class: https://vhbio.com/contact/.
